Summary
The most commonly used cell-based assays for measuring cytotoxicity often show poor reproducibility, require 24 h to complete, and rely on a nonspecific response, namely cell death. Better assay technologies are required, both for drug and environmental toxin screening, and for applications in the field. An example of a public health and safety need is the determination of the presence of protein toxins such as the plant toxin ricin. We describe an assay for ricin that provides a response that is relevant to the mechanism of ricin activity and permits a much faster readout than commonly used assays for cytotoxicity. The assay relies on the response of an engineered reporter cell line that was produced by stably transfecting Vero cells to express green fluorescent protein (GFP) under the control of a cytomegalovirus (CMV) promoter. Unlike the other assays, monitoring cellular GFP on a per-cell basis allows detection of the effect of the toxin on cell activity before significant cell death occurs, and provides a response that is relevant to the physiological function of the toxin.
Description
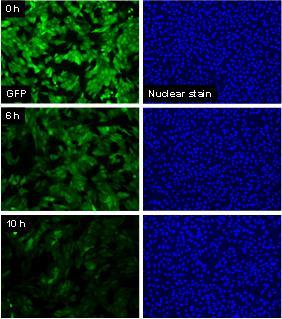
Ricin is a type 2 ribosome inactivating protein (RIP) belonging to the A-B family of toxins and is of particular interest because of its potential use as a biological weapon and the dearth of rapid and sensitive assays that could assist detection and decontamination. The ricin toxin inhibits protein synthesis by first binding to cell surface galactose residues, then entering the cell via endocytosis and inactivating ribosomes catalytically and irreversibly. A convenient, cell-based assay that was specific to the action of such a toxin would allow the biological activity of the protein to be assessed, since it would report the sensitivity of the cell to the receptor mediated endocytosis and cytoplasmic transport of the toxin, as well as the ultimate effect on protein synthesis. In this study, we used a stably transfected line of Vero cells that expresses green fluorescent protein (GFP), and we measured the loss of cellular GFP as an indicator for protein synthesis inhibition by the ribosome inactivator, ricin. The GFP construct used here consists of the sequence for dsEGFP (destabilized enhanced GFP) ligated to the promoter sequence for cytomegalovirus (CMV). Under control of the CMV promoter, GFP is constitutively expressed by the cells, and because it contains a proteasome-targeting peptide sequence (PEST), it is rapidly degraded with a half life of approximately 2-3 h as we have directly demonstrated by examining degradation rates in individual cells (Halter et al., Cytometry Part A, 2007). Changes in the synthesis rate of GFP due to interference with ribosome activity has a rapid effect on decreasing the cellular GFP levels in affected cells.
We compared this assay, which is based on quantitative imaging to measure GFP levels in cells over time, with the MTT assay, the MultiTox-Fluor Multiplex Cytotoxicity Assay, and the CellTiter-Glo Luminescent Cell Viability Assay, all of which are based on measurements of an overall response from a well or plate of cells. These whole-well assays are intended to report cellular enzyme activity, but their results are convoluted by the number of cells present. The GFP assay, on the other hand, is more specific to the mechanism of action of the toxin, is a more rapid indicator of toxin activity, and allows quantification of the fluorescence intensity of cellular GFP using quantitative fluorescence imaging of individual cells. A DNA counterstain is used to monitor the number of cells in each well, so that the total GFP intensity can be normalized to cell number.
Major Accomplishments
- The GFP-based assay is specific for ribosome inhibition, providing a mechanistically-based assay for cytotoxicity.
- The GFP-based assay described here is highly reproducible, requires no added reagents, and is more rapid than conventional cytotoxicity assays such as MTT, MultiTox-Fluor Multiplex Cytotoxicity assay, and CellTiter-Glo Luminescent Cell Viability assay.
- The effect of ricin on GFP intensity occurs before significant cell death has occurred (approximately 6 h), so the measurement is not confounded by a significant reduction in cell number.
- Many live/dead cell assays can be confounded by cell activity expressed during the apoptotic process. In order to attain accurate measurements, these assays must be performed after cells die and detach from the plate (24 h – 48 h after intoxication). Measuring individual cell translation inhibition can therefore be done earlier (6 h) with more accurate measurements.
- The GFP-based assay provides a limit of detection to ricin of less than 1 ng/ml (p=0.02) in just 6h.
M. Halter, A. Tona, K. Bhadriraju, A. L. Plant, J. T. Elliott, "Automated live cell imaging of green fluorescent protein degradation in individual fibroblasts." Cytometry A. 2007 Oct;71(10): 827-34.

