Anne L Plant (Assoc)
NIST Fellow
Research Activities
While much of the study of the life sciences has been empirical in nature, the field continues to evolve into a science that is founded on fundamental physical principles. Understanding how physical principles apply to biological systems will allow predictive understanding of these complex processes. In addition, better fundamental understanding will help to make the manufacturing of advanced therapy products more efficient and reliable.
Our current research focuses on single-cell measurements from live cell microscopy, and development of theoretical approaches that lead to predictive understanding of complex cellular systems. Currently, we are examining small molecular networks in engineered induced pluripotent stem cells with a statistical thermodynamics model. The heterogeneity of cellular phenotypes within isogenic populations provides a window into the diversity of ways the cellular machinery can operate to process information. This diversity of expression reflects the stochastic fluctuations that occur as connected network variables interact with one another to effect up- and down-regulation.
Our goal is to measure the kinetic constants for those fluctuations, and the correlations between them, to provide a thermodynamic basis for understanding which cellular measurands are the most important for controlling a particular phenotype. This work is motivated by a critical challenge in characterizing cell-based therapies, namely what should be measured that will provide predictive information about the cell product to assure desirable functionality. Carrying out this study involves a large number of NIST collaborators to address challenges in rapid non-perturbing quantitative imaging, CRISPR-Cas genome editing of reporter iPSC lines, image analysis including the use of CNN inference, and theoretical modeling.
Key words: Cell therapies, Regenerative Medicine, Pluripotent stem cells, Fluorescence microscopy, Live cell imaging, Image analysis, statistical thermodynamics
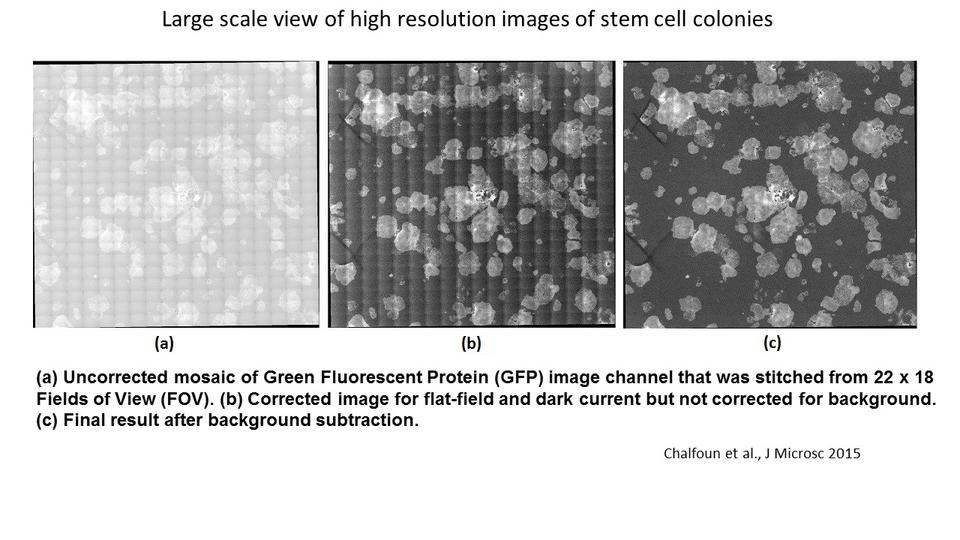
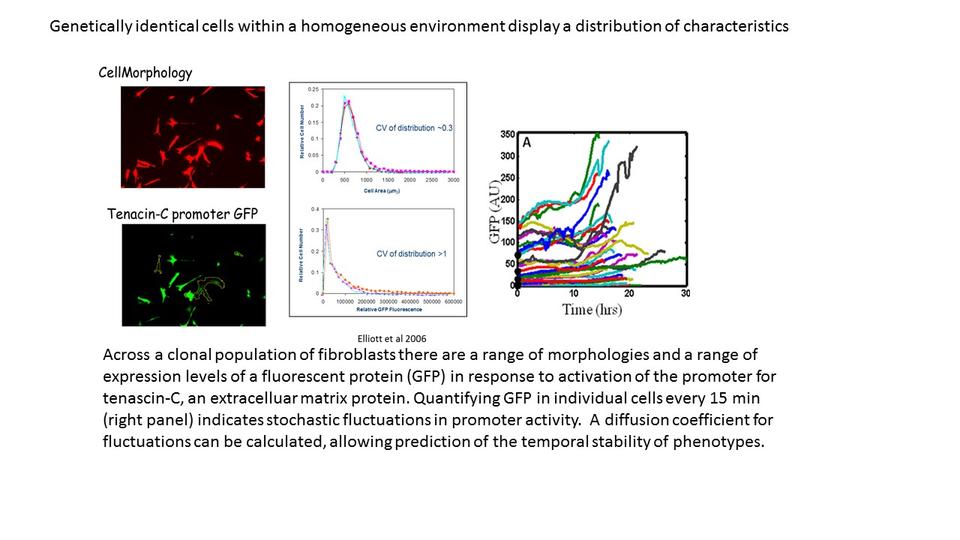
Selected Publications
Hubbard JB, Halter M, Sarkar S, Plant AL (2020) The role of fluctuations in determining cellular network thermodynamics.PLOS ONE 15(3): e0230076.
Sisan D.R., Halter M., Hubbard J.B., Plant A.L. (2012) Predicting rates of cell state change due to stochastic fluctuations using a data-driven landscape model. PNAS 109, 19262-19267
Michael Halter, Steven Lund, Adele Peskin, Ya-Shian Li-Baboud, Peter Bajcsy, Oleg Aulov, Daniel J. Hoeppner, Josh G. Chenoweth, Suel-Kee Kim, Ronald D. McKay, Anne L. Plant. (2019) A Latent Variable Model for Evaluation of Disparate Ratings of Stem Cell Colonies by Two Experts. BioRxiv doi: https://doi.org/10.1101/746057
Keating, S.M, D. Lansing Taylor, Anne L. Plant, E. David Litwack, Peter Kuhn, Emily J. Greenspan, Christopher M. Hartshorn, Caroline C. Sigman, Gary J. Kelloff, David D. Chang, Gregory Friberg, Jerry S. H. Lee, and Keisuke Kuida (2018) Opportunities and Challenges in Implementation of Multiparameter Single Cell Analysis Platforms for Clinical Translation. Clin Transl Sci doi:10.1111/cts.12536
Peskin, Adele P., Lund, Steven P.; Li-Baboud, YaShian; Halter, Michael W.; Plant, Anne L.; Bajcsy, Peter. (2014) Automated Ranking of Stem Cell Colonies by Translating Biological Rules to Computational Models. 5th ACM Conference on Bioinformatics, Computational Biology and Health Informatics.
Halter, Michael W. Bier, Elianna; DeRose, Paul C.; Cooksey, Gregory A.; Choquette, Steven J.; Plant, Anne L.; Elliott Jr., (2014) An automated protocol for performance benchmarking a widefield fluorescence microscope. Cytometry. doi: 10.1002/cyto.a.22519.
Bhadriraju, Kiran, Michael Halter, Julien Amelot, Peter Bajcsy, Joe Chalfoun, Antoine Vandecreme, Barbara S. Mallon, Kye-yoon Park, Subhash Sista, John T. Elliott, Anne L. Plant (2016) Large-scale time-lapse microscopy of Oct4 expression in human embryonic stem cell colonies. Stem Cell Research, 17:122 doi:10.1016/j.scr.2016.05.012
Peterson, Alexander, Michael Halter, Anne Plant, and John Elliott (2016) Surface plasmon resonance microscopy: achieving a quantitative optical response. Review of Scientific Instruments, 87:9 DOI: 10.1063/1.4962034
Peterson, Alexander W.; Halter, Michael W.; Tona, Alessandro; Plant, Anne L. (2014) High Resolution Surface Plasmon Resonance Imaging of Cells. BMC Cell Biology. 15:35 DOI: 10.1186/1471-2121-15-35
Dima, Alden, John Elliott, James Filliben, Michael Halter, Adele Peskin, Javier Bernal, Marcin Kociolek, Mary Brady, Hai Tang, Anne Plant. (2011) Comparison of segmentation algorithms for fluorescence microscopy images of cells. Cytometry Part A. 79A: 545-559.
Halter, Michael , Daniel R. Sisan, Joe Chalfoun, Benjamin L. Stottrup, Antonio Cardone, Alden Dima, Alessandro Tona, Anne L. Plant, John T. Elliott. (2011) Cell cycle dependent TN-C promoter activity determined by live cell imaging. Cytometry Part A. 3A: 192-202.
Bhadriraju, K., Chung, K.-H., Spurlin, T. , Haynes, R., Elliott, J.T., Plant, A.L. (2009) Downregulation of Focal Adhesion Kinase in vascular smooth muscle cells on Type I collagen is associated with DDR2 activation independent of collagen matrix mechanical properties. Biomaterials 30(29):5486-96.
Halter, M., A.L. Plant, J.B. Hubbard, A.Tona and J.T. Elliott. (2009) Cell Volume Distributions Reveal Growth Rates and Division Times. Journal of Theoretical Biology 257, 124

