Summary
The National Institute of Standards and Technology is developing a peptide mass spectral library as an extension of the NIST/EPA/NIH Mass Spectral Library. The purpose of the library is to provide peptide reference data for laboratories using mass spectrometry to discover disease-related "biomarkers." Using mass spectral libraries to identify these compounds is more sensitive and robust than interpreting the mass spectra by theoretical methods. These databases are freely available for testing and development of new applications (http://peptide.nist.gov).
Description
Intended Impact
Modern mass spectrometers used in the field of proteomics are capable of profiling thousands of peptides in a single experiment. Each of these peptides is fragmented to form a mass spectrum. Therefore, interpretation of these mass spectra is a critical step in the experimental workflow. Since peptide mass spectra represent physical properties of these molecules, standard interpretation of these mass spectra has the potential to improve the success rate of all discovery experiments in proteomics.
Objective
Biological mass spectrometry is a critical tool in the search for new markers of disease. These markers will be used as targets for tomorrow's diagnostics and therapeutics NIST researchers are using their expertise in building mass spectral libraries for other small molecules to compile a comprehensive library of consensus peptide mass spectra from human samples and other important model organisms. Developing a standard method for interpreting these mass spectral data is critical for establishing and advancing this technology.
Goals
- Develop a mass spectral library of all biologically relevant peptide ions
- Provide the library in a form that is easily searchable using software tools
- Update and maintain the library as a reference data resource
Research Activities and Technical Approach
While the mass spectrometers used to identify peptides in proteomics have improved greatly over that past ten years, computer algorithms for peptide identification have not. Traditionally, this process involves a step wherein theoretical peptide fragmentation spectra are predicted from protein sequences. These spectra typically contain peaks at the correct m/z values but contain little or no information about their relative intensities (i.e. peak heights) or less common fragmentation products. Mass spectral libraries capture this information from measurements, enabling the use of more sensitive search algorithms. The use of these algorithms and libraries (1) will lead to a higher percentage of identified spectra at the same level of reliability, and (2) will greatly increase the robustness of the peptide identification step.
The data for this project is both being generated 'in-house' at NIST and collected from many outside sources. NIST also has data exchange agreements with several international proteomics data repositories in order to efficiently share the most relevant data.
To date, several of the libraries, including human, yeast and E. coli, represent significant coverage of the proteomes and are suitable for routine uses.
Their use, in combination or as an alternative to sequence-based identification methods, has been shown to double the number of peptide identifications for some data sets.
Major Accomplishments
- Large peptide mass spectral libraries have been developed for human and several model species
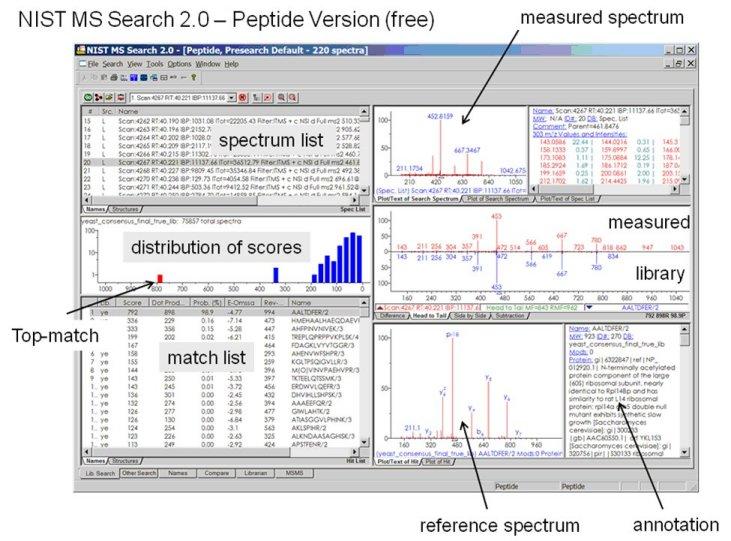
- A peptide version of NIST MS Search 2.0 is available (Fig. below)
- A website has been launched for distribution of the libraries and software (http://peptide.nist.gov)
Publications
- Development and Validation of a Spectral Library Searching Method for Peptide Identification from MS/MS, Lam, H., Deutsch, E.W., Eddes, J.S., Eng, J.K., King, N., Stein, S.E., Aebersold, R. Proteomics, 2007 Mar., 7(5), pp. 655-657.
- Building Consensus Spectral Libraries for Peptide Identification in Proteomics, Lam, H., Deutsch, E.W., Eddes, J.S., Eng, J.K., Stein, S.E., Aebersold, R. Nat. Methods, 2008 Oct, 5(10), pp. 873-875.

