Summary
The National Institute of Standards and Technology is responsible for providing an evaluated tandem mass spectral library for reliable compound identification. The principal use of the library is for the identification of compounds by matching fragmentation “fingerprints” (mass spectra) of their ions as generated by electrospray ionization and atmospheric pressure chemical ionization. In conjunction with separation by liquid chromatography, this technique is widely used to identify the chemical components of complex fluids and tissues derived from substances of great practical interest, including disease-related metabolites, drugs, foods, environmental contaminants and many more. This database is available from distributors for integration with existing instrumentation (http://chemdata.nist.gov/mass-spc/msms-search/). NIST23 contains 60% more compounds than NIST20.
Description
Intended Impact
Modern mass spectrometers commonly generate ions by electrospray mass spectrometry which are separated and then fragmented by collisional dissociation. This process yields fragment ion ‘fingerprints’ (or mass spectra) that are widely used to identify the individual components in a complex mixture. In this way, many hundreds of different compounds over wide ranges of concentrations can be fragmented in a single experiment. These ‘tandem’ mass spectra can serve to identify these chemicals, which is a critical and error-prone step in the experimental workflow. Since mass spectra reflect physical properties of these molecules, they can provide reproducible ‘fingerprints’, enabling the reliable identification from their mass spectra. By providing high-quality spectra over the range of conditions employed by mass spectrometers, the library can greatly improve the success rate of confident chemical identification – a necessary step in the molecular analysis of any chemical or biological material.
Objective
The objective of this program is to provide a practical means of reliably identifying chemical components in biological and other complex mixtures through their mass spectral ‘fingerprints’ stored in a NIST database. These compounds include human and plant metabolites, pollutants, contaminant, drugs and a wide range other societally important compounds.
Goals
- Develop a comprehensive mass spectral library of compounds of practical interest, especially those of biologically relevance
- Provide the library in a form that enables the reliable identification of compounds using efficient searching software tools
- Update and maintain the library as a reference data resource
Research Activities and Technical Approach
The mass spectrometers used to identify metabolites and other small molecules have been improved greatly over the past ten years. While hundreds of compounds may be detected in a single experiment, identification of these compounds depends on the availability of libraries of standard reference mass spectra of all those compounds. Mass spectral libraries are built from measured spectra of known compounds and enable the use of sensitive search algorithms. The use of these algorithms and libraries (1) will lead to a higher percentage of identified spectra at a high level of reliability and (2) will greatly increase the robustness of the metabolite identification step.
To date, this mass spectral library contains 2,374,064 spectra for 399,267 precursor ions of 51,501 compounds of biological and environmental relevance (including metabolites, drugs, pesticides, bioactive peptides, amino acids and small peptides, lipids, phospholipids and glycolipids, sugars and glycans, extractable and leachable compounds, surfactants, and various contaminants). The spectra in the libraries have all been experimentally validated, critically evaluated, and annotated in great detail.
Major Accomplishments
- A large metabolite mass spectral library has been developed
- A new, chemical structure based method for continued library development has been implemented
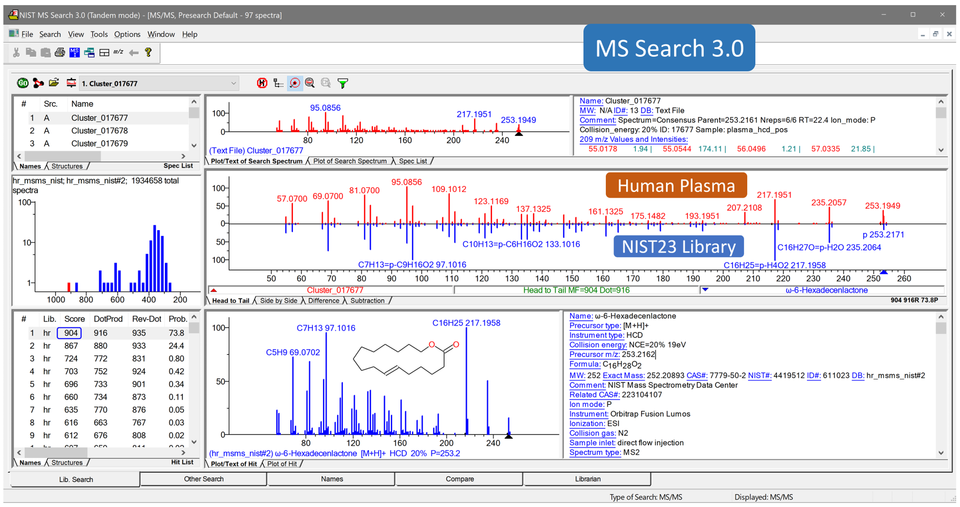
- A new version of NIST MS Search 3.0 is available (Fig. below)
- A website has been updated for distribution of the libraries and software (http://chemdata.nist.gov/mass-spc/msms-search)
Publications
- Extending a Tandem Mass Spectral Library to Include MS2 Spectra of Fragment Ions Produced In-Source and MSn Spectra. Xiaoyu Yang, Pedatsur Neta and Stephen E. Stein; Journal of The American Society for Mass Spectrometry, November 2017, Volume 28, Issue 11, pp 2280–2287, DOI: 10.1007/s13361-017-1748-2
- Quality Control for Building Libraries from Electrospray Ionization Tandem Mass Spectra. Xiaoyu Yang*, Pedatsur Neta and Stephen E. Stein; Anal Chem. 2014 Jul 1;86(13):6393-400. doi: 10.1021/ac500711m. Epub 2014 Jun 13. DOI: 10.1021/ac500711m