Summary
Induced pluripotent stem cell (iPSC) populations are complex, dynamic and heterogeneous. Individual cells within a population are constantly changing, while maintaining the capacity to differentiate in numerous possible cell types. Sophisticated measurement tools are required to adequately describe and develop predictive models for complex cellular systems such as these. The technology to record live cell images from cellular populations has been available for some time, but only recently has it become routine to derive quantitative data from these image sets using image analysis. We have focused on developing live cell imaging tools to monitor large numbers of single cells and to quantify changes in morphology and gene expression using fluorescence protein reporters.
Description
As part of our program in Cell Biology, our live cell image program supports the advancement of induced pluripotent stem cell (iPSC) technology in three ways:
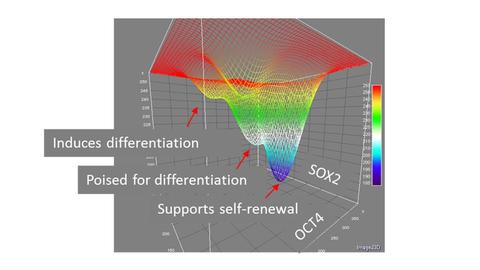
1) Identification of process control measurements: A critical component to the translation of iPSCs into therapeutic applications is to design principles for predictably and reproducibly culturing cells and efficiently differentiating them into cell types of interest. Live cell imaging provides ‘high-resolution’ measurements in the sense that we collect time-dependent data from large numbers of individual cells, such as morphologies, cell divisions, and expression of fluorophore reporters of transcription factors. We can then use these data to discover lower resolution measurements, such as the activity of a small number of biomarkers at some frequency that can serve as critical metrics to guide process control during manufacturing of pluripotent stem cells.
2) Interpreting biomarkers: Cells are stochastic and dynamic and the expression of biomarkers can change over time. The predictive power of a biomarker or a set of biomarkers may be inferred by examining the history of that cell by tracking forward and backward in time through a time lapse image set.
3) Predictive modeling: We have shown that fluctuations in promoter activity can be used in combination with appropriate models to predict rates of state change in cell populations. Similar mathematical models that can inform bioprocessing decisions during scale-up will be critical to obtaining iPSC populations with a desired set of characteristics.
Over the past several years, we have developed tools for measuring parameters related to size, shape, and intensity from single cells over time (Halter Cytometry 2011). We have also developed modeling tools for using the temporal information to model the stochastic and deterministic components of gene expression (Sisan PNAS 2012; Lund Phys Chem B 2014; Hubbard 2020).