Summary
The natural world is a magnificent place with observable, via measurement and inference, physical structures spanning at least 43 orders of magnitude in space and in time. Life itself spans a broad range of these spatial and temporal scales, leaving an intricate mystery: How does the molecular machinery give rise to it all? How do processes all the way down to the atomic scale, 10-10 m and 10-15 s, lead to individual living structures all the way up to 103 m and 1011 s. Moreover, living processes conceal their wonder in a shroud of complexity, often making mechanistic processes difficult to directly measure or even infer. The goal of the program in molecular physiology is to design measurements, data analyses, and theoretical frameworks that capture biomolecular processes from the atomic scale (10-10 m, 10-15 s) to the cellular scale (10-6 m, 103 s; not including, e.g., Paramecium tetraurelia or Caulerpa taxifolia).
Description
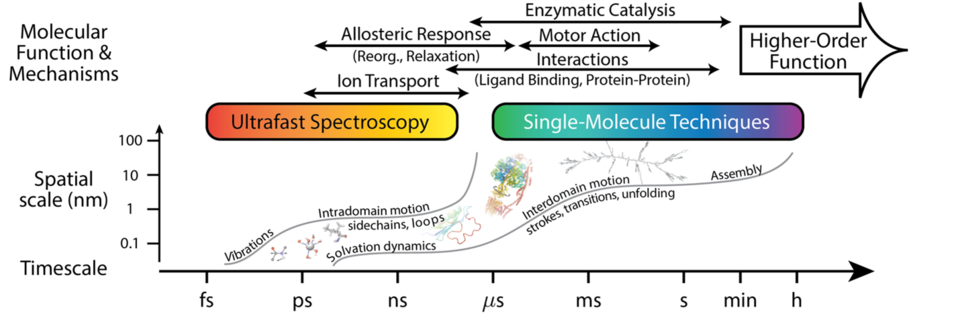
In close collaboration with experimentalists, we are seeking to increase the resolution, scope, and throughput of single-biomolecule and ensemble techniques, such as nanopore-based biomolecular analysis, ultrafast vibrational spectroscopy, and FRET, among others. These developments rely heavily on ability to fabricate and employ novel nanostructures, both bottom up and top down, to enhance specificity and decrease uncertainty. Advances in these techniques are proceeding in concert with developing accurate theoretical and computational methods, including machine learning and (all-atom) molecular simulation, as well as with approaches for comprehensive experimental data analysis. The result—high quality data and interpretation—enables biophysical and biological researchers to distill general principles and design critical tests of biomolecular operation across scales, see the figure.
We have several recent and ongoing developments:
- Ultrafast vibrational spectroscopy: While ubiquitous, energy redistribution remains a poorly understood facet of the nonequilibrium thermodynamics of biomolecules. At the molecular level, finite-size effects, pronounced nonlinearities, and ballistic processes conspire to produce behavior that diverges from the macroscale. We have shown that transient thermal transport, such as that measured by ultrafast vibrational spectroscopy, reflects macromolecular energy landscape architecture through both (i) the topological characteristics of the conformational ensemble and (ii) the nonlinear processes that mediate dynamics. While the former determines transport pathways via molecular contacts, the latter reflects the ruggedness of the landscape for local motion of atoms and molecular fragments. Unlike transport through small-molecule systems, such as alkanes, nonlinearity dominates over coherent processes at even quite short time- and length-scales. Our exhaustive all-atom simulations and novel local-in-time and space analysis, applicable to both theory and experiment, permit dissection of energy migration in biomolecules. The approach demonstrates that vibrational energy transport can probe otherwise inaccessible aspects of macromolecular dynamics and interactions that underly biological function.
- FRET: Understanding the folding process of DNA origami is a critical to the broader implementation of nucleic acid nanofabrication technology but is notably non-trivial. A DNA origami structure is formed by several hundred cooperative hybridization events – folds – between spatially separate domains of a scaffold, derived from a viral genome, and oligomeric staples, each of which is difficult to detect independently. We characterized the unit process of DNA origami folding under highly controlled conditions, measuring yield as a function of folding distance and staple/scaffold ratio via a carefully designed fluorescent reporting system. By examining a single fold, and thus limiting the number of possible topology states, we enable evaluation of a thermodynamic model for this unit-step system. This approach to the folding problem elucidates a predicted but previously unobserved blocked state that acts as a limit on yield for single folds, which may manifest as a barrier in whole origami assembly.
- Electrophysiology and transport measurements: Biological ion channels evolved to have high transport rates and high selectivity. Determining and quantifying the transport mechanisms responsible for this feat is paramount to understanding biological systems. We are developing a landscape theory of optimization of ion channels, both biological and synthetic. Interactions between translocating ions and the channel occur at subnanometer distances. This entails that there are large electrostatic interactions and dehydration energies, the balance of which determines selectivity and rates. Importantly, the susceptibility of transport and selectivity to minute changes in distances – changes on the order of picometers – is enormous. Biological systems can exploit this susceptibility via variations in protein structure that steers the local electrostatic and steric conditions. We have demonstrated how this works in a synthetic selectivity filter and proposed how to experimentally probe this system, as well as examined the exemplary KcsA channel. The landscape theory presents a new paradigm that will enable pinpointing the functional features in biomolecular operation.
Work with us
Please contact Michael Zwolak at mpz [at] nist.gov (mpz[at]nist[dot]gov) to inquire about postdoctoral and collaborative opportunities.
Postdoctoral opportunities are available through a National Research Council (NRC) Fellowship, a 2-year program with application deadlines of February 1 and August 1 each year.
Additional information about the specific opportunities and the program are below:
- Research opportunity in Theoretical Nanoscale Biophysics
- NIST NRC Research Associateship Program Information
Selected Publications
- Elenewski, J. E., Velizhanin, K. A., & Zwolak, M., Topology, landscapes, and biomolecular energy transport, Nat. Comm. 10, 4662 (2019).
- Sahu, S., Elenewski, J. E., Rohmann, C., & Zwolak, M., Optimal transport and colossal ionic mechano-conductance in graphene crown ethers, Sci. Adv. 5, eaaw5478 (2019).
- Sahu, S. & Zwolak, M., Colloquium: Ionic phenomena in nanoscale pores through 2D materials, Rev. Mod. Phys. 91, 021004 (2019).
- Sahu, S., Di Ventra, M., & Zwolak, M., Dehydration as a universal mechanism for ion selectivity in graphene and other atomically thin pores, Nano Lett. 17, 4719 (2017).
- Sahu, S. & Zwolak, M., Ionic selectivity and filtration from fragmented dehydration in multilayer graphene nanopores, Nanoscale 9, 11424 (2017).

