Summary
Monoclonal antibody (mAb) therapeutics generate $75 billion dollars in revenue for biopharmaceutical companies and constitute nearly 50 % of top therapeutics by sales (2019 data). They are required components of any immunoassay measurement device/technique, from COVID-19 rapid-tests to biomarker screening assays. Designing and modifying mAbs to control their function is an active area of research and development. However, because of their size (> 10 nm), our ability to measure solution-state dynamics of mAbs and link those dynamics to important functional attributes is limited.
Description
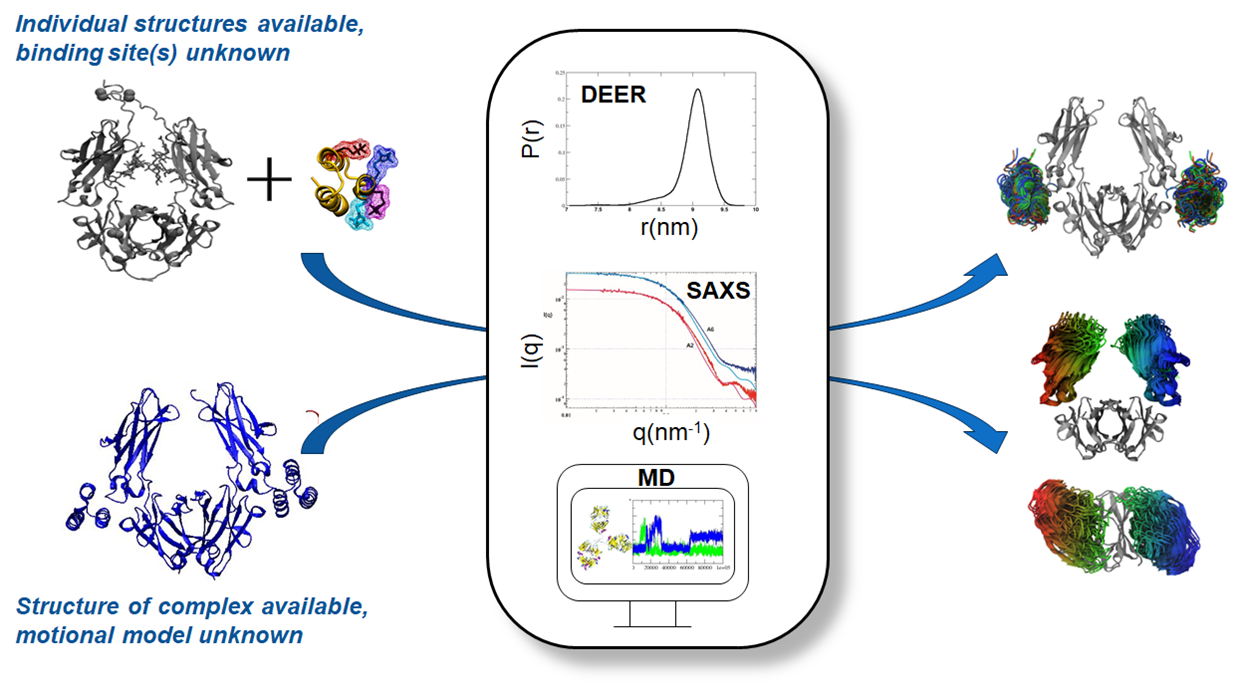
Through collaboration with researchers at the Institute for Biotechnology and Bioscience Research (IBBR), we have created a library of site-specific spin-labeled and stable isotope-labeled binding partner proteins for the NIST monoclonal antibody (NISTmAb) reference material (RM8671). Using pulsed dipolar EPR spectroscopy, we have determined point-to-point interspin distance distributions in a collection of spin-labeled protein/NISTmAb complexes. Combining molecular dynamics (MD) simulations, small-angle X-ray scattering (SAXS) and double electron-electron resonance (DEER) measurements, we have developed a model of the dynamics of the cellular receptor-binding portion (Fc) of the NISTmAb in solution, information which had been previously unavailable yet needed to develop testable hypotheses about how/whether solution-state dynamics control mAb functional properties (e.g., solution viscosity of mAb formulations, aggregation propensity, cross reactivity). Because the isotopically-labeled proteins generated for this project are compatible with any mAb in the immunoglobin G class, they provide a robust and general approach for obtaining structural & dynamics information on mAbs used by industrial and academic stakeholders.
Publications/Presentations
Structure and dynamics of flexibly-linked, multi-domain proteins determined using spins, scattering, and simulations, Szalai, V.A. Bergonzo, C., Grishaev, A., Schmidt, T., Southeastern Magnetic Resonance Conference, Atlanta, GA, October 25, 2024.
Structure and dynamics of flexibly-linked, multi-domain proteins determined using spins, scattering, and simulations, Szalai, V.A. (invited) Bergonzo, C., Grishaev, A., Schmidt, T., International Virtual Electron Paramagnetic Resonance Meeting, September 25, 2024.
Structure and dynamics of flexibly-linked, multi-domain proteins determined using spins, scattering, and simulations, Szalai, V.A. Bergonzo, C., Grishaev, A., Schmidt, T., Rocky Mountain EPR Conference, Copper Mountain, CO, August 7, 2024
Integrated analysis of inter-electron spin distances, X-ray scattering, and molecular simulation to resolve the dynamic structure of flexibly-linked multidomain biologics, Szalai, V.A. Bergonzo, C., Grishaev, A., Schmidt, T., Alpha-Twists and Beta-Turns of Recombinant Protein Production, National Cancer Institute, Frederick, MD March 21-22, 2024

