File Rule
Basics
Trajectories perform 3 core tasks:
- Visit a series of instrument states. Each state can be comprised of motor positions, temperatures, etc.
- Perform a count at each state
- Store the results
Trajectories allow you to configure how step 3 (store the results) is performed. In particular:
- When data files are created.
- How data files are named.
- How data is arranged within/across data files.
Additionally, NICE can write data to multiple formats simultaneously. Two common formats are:
- column – A tabular text format.
- NEXUS – A common neutron scatter format for organizing data. NICE uses the Hierarchical Data Format (HDF) binary flavor.
NOTE: Trajectories do not allow you to configure active writers (which data formats are written), but this is changeable and is described here: Writers.
Each data format is governed by a “writer” (a small python program) which is responsible for the details of how data is written. Users can describe how they want writers to store a trajectory’s data by providing “hints” in the form of “file rules” (specified by setting special variables). For example, a user could provide a hint by setting the variable fileName = “myfile”, which would result in the NEXUS writer producing the myfile.nxs.<INT> file and the column writer producing the myfile.<INT>. file, where <INT> represents a unique 3-letter instrument tag.
NOTE: Setting filename directly is NOT a wise idea. This is explained in more detail in Example 1.
At various times, while a trajectory is executing each point (move + count) it broadcasts the instrument state, counts and sends trajectory variables to each active writer. Unlike other trajectory variables, file rules are expressions, which are defined once and automatically re-evaluated per point, before data is sent to each writer. Understanding available file rule variables and using more complex formulas will allow you to arrange and distribute data across files in any desired manner. For simplicity, we will focus solely on the output of the NEXUS writer in the examples below, but the concepts discussed extend to all writers/formats.
Example 1
Here is a trajectory which visits the following temperatures: [100,125,150,175,200]. At each iteration, the trajectory goes through two polarization states [UP, DOWN] for a total of 10 points. At each point, we will count for fixed period of 5 seconds.
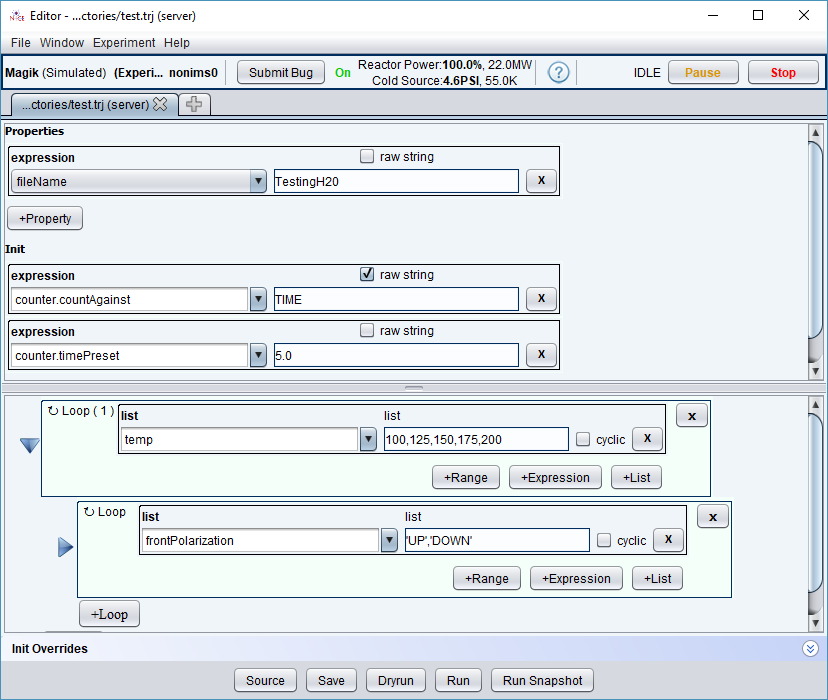
We set the fileName expression to be “TestingH20”, which evaluates to the fixed value “TestingH20” at every point. Not surprisingly, all data points are written to one data file named “TestingH20”:

While this looks like a reasonable scheme, if you rerun the same trajectory, fileName will still evaluate to the same “TestingH20” name, causing the writers to deal with possible name collisions.
To improve this scheme, we can change the fileName expression to be:
fileName = “TestingH20” + fileNum
where fileNum is a special variable that can be attached to the end of the file name. It automatically increments every time a trajectory is run.
Every time we run the same trajectory, a data file will be generated with a new name: the provided name “TestingH20” followed by an incrementing integer:
TestingH201.nxs
TestingH202.nxs
TestingH20<i>.nxs
After all this, it turns out you’ll get similar behavior if you simply don’t use fileName at all! The reason is that NICE provides several defaults:
- fileName = filePrefix + fileNum
- filePrefix = trajName
- trajName = <the base name of the trajectory file being run>
NOTE: Technically fileName = "String(filePrefix) + String(fileNum)" which effectively concatenates filePrefix and fileNum (formally defaults to "sprintf('%s%d',filePrefix,fileNum)").
So, if you don’t specify any rules whatsoever, running the same trajectory (stored as test.trj) would generate the following data files:
test1.nxs
test2.nxs
test<i>.nxs
NOTE: It’s at discretion of each writer to decide what to do about file name collision. The NEXUS writer avoids collisions by modifying the original file name; It appends character sequence <_Ai> where i represents an increment of how many times the file with this name has been stored on disk.
Example:
TestingH20.nxs
TestingH20_A1.nxs
TestingH20_A2.nxs
Other writers might append data to the same file, so it’s wise to not rely on this.
Example 2
Let’s now concentrate on how we can organize data within a running trajectory. Let’s reuse our original trajectory from Example 1, which visits the following temperatures [100,125,150,175,200] and at each iteration goes through two polarization states [UP, DOWN], for a total of 10 points.
What if a user wants each measurement to go into a separate file based on the temperature value? This can be accomplished by setting filePrefix to the following expression:
filePrefix = "temp_" + temp + "_"
If we assume fileNum had reached 4 before the trajectory was run, then fileNum would be incremented to 5. The output would include 5 different files based on temp value.

It’s important to note that our files were named using the desired value temp, as evaluated in the trajectory, and NOT the value of temperature that was achieved. This is generally preferred as it results in consistent and predictable file names.
It’s also important to note that even though device and node names are not case sensitive, variable names in trajectories are. When users manipulate the “temp” device in a trajectory, that name essentially becomes a variable in the JavaScript engine. JavaScript is case-sensitive, and users must use consistent capitalization, or the results will be unpredictable.
Example 3
The NEXUS writer allows for storage of several scans in the same file as separate entries defined by entryName. If unspecified, then entryName defaults to an empty string in the editor. The writer would then store all the data under a default entry named "entry".
Let’s redefine the above trajectory to store the points in five files, with each file containing two entries based on the polarization state: “UP” or “DOWN”.
Here is an example of the desired NEXUS hierarchy for one file:
- temp_150_5.nxs
- DOWN
- UP
Here is how the expressions would be set up to achieve this:

filePrefix = "temp_" + temp + "_"
entryName= frontPolarization
NOTE: While inspired by NEXUS, all writers can observe the entryName rule and it is at each writer’s discretion what is done with it. Some writers may use the rule, while others may ignore it. Please see the section on Writers for more details.
Example 4
As demonstrated in the previous example, a trajectory can move a node like temp through a series of desired values and further use these desired values in formulas and file expressions. However, what if we want to use a node’s current value in the file name? We cannot do that because trajectories cannot access the “live” value of a node as it changes during the trajectory’s execution. What is available to trajectories is a snapshot of initial node states, as they were, right before the trajectory started execution. These values can be accessed via the start prefix (formally known as live) followed by the full node name (for example: start.lakeshore340.primaryNode).
Let’s revisit the trajectory we’ve been using in the past several examples and make a few changes. Firstly, let’s name our files based on the sample we’re using, rather than simply making it “temp”. This assumes the current sample has already been configured and has a been given a name (the current sample’s name is stored in the node sample.name).
We’ll change the rule for filePrefix to be:
filePrefix = start.sample.name + "_temp_" + temp + "_"

Example 5
In the previous examples we wanted to put data associated with each distinct temperature into their own files. To do this, we made fileName a function of temperature. This worked for those examples, but what if the desired temperatures were long, decimal values? What if many changing states, like magnet fields or motor positions, were combined to create a file name? We can imagine ending up with an awkward file name like:
myFile_temp_127.125_mag_4.0625_motorX_20.45.nxs.<INT>
So, the question is: how can we group data, based on common states like temperature, but not be forced to include temperature in the name of a file?
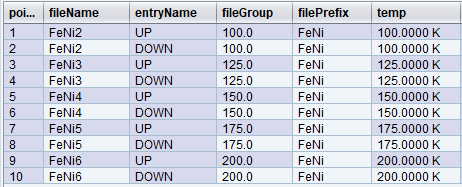
The answer is to use the fileGroup expression. At every point, the trajectory engine will check if fileGroup has changed and, if so, will increment fileNum. Otherwise, the trajectory engine will pick the fileNum associated with the existing fileGroup. By default, fileGroup is empty – and in all the examples above, fileGroup was always empty and unchanging, and so fileNum was only incremented once per trajectory.
Let’s change our original trajectory a bit more. In this example, we decided to visit the following temperatures: [100,125,150,175,100,200] and at each iteration go through two polarization states [UP, DOWN] for a total of 12 points.
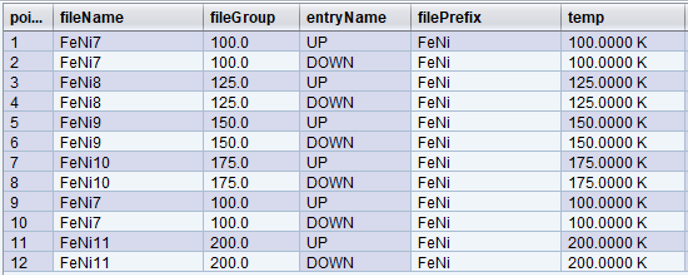
Since fileName now depends on fileGroup, notice that the second time temperature reached 100.0 K, at point 9, the data was written to the same data file as in point 1, [FeNi7]. Also, when a new fileGroup was added for 200.0K at point 11, the fileNum incremented again, hence the fileName, [FeNi11].
To produce similar output to that above, we can use the following file rules:
filePrefix = start.sample.name
entryName= frontPolarization
fileGroup = temp
The exact names of files will depend on the initial value of the fileNum variable.
Example 6
If the users are interested in manipulating several samples in the same trajectory and record corresponding sample information in the file name then they would have to follow special rules. We have changed how samples are stored on the server, they are no longer represented as a device, but as a table stored in persistent configuration. As a result trajectories need to handle this case separately. Imagine a trajectory that goes through several samples and at each sample changes the temperature.
loop sampleId (1,2,3)
loop temp (300,350,400)
Also lets assume that the user wants data for each sample to be stored in a separate data file. In other words, we will have three files named after each sample name with three data points in it.
Important node: before each trajectory runs, it stores almost all of the instrument state in the variable called start (see example 4 for details). The start variable also incudes the entire sample table. It can be accessed by calling start.sampleTable. For example, sample 1 info can be accessed by calling start.sampleTable(1) followed by the name of the property. Hence, we can describe file name by the following rule:
filePrefix = "start.sampleTable.get(parseInt(sampleId)).get('name')"
Here is the corresponding dryrun result:

Appendix
File Variables
These are special, read-only keywords maintained by the trajectory engine, which can be used to construct file expressions.
fileNum
- represents a counter; every time a point is about to count, it checks if a new fileGroup is available and increments fileNum.
- While the trajectory is running, if a fileGroup never changes, then the counter only increments once.
- It resets back to zero for every new experiment.
instFileNum
- the same as fileNum, except that it does not reset for every new experiment.
- In practice this should increment forever.
pointNum
- starts at zero for each new trajectory and increments once per point.
expPointNum
- starts at zero for each new experiment and increments once per point.
- Switching to an old experiment will not reset expPointNum.
File Expressions
fileName
- is the final value used by writers to construct file names.
- Default: "String(filePrefix) + String(fileNum)"
- If overwritten, this can cause name collisions when the same trajectory runs again.
- Can be specified per instrument to be displayed in the editor.
filePrefix
- can be specified instead of fileName to guarantee uniqueness of the data files.
- Default: filePrefix = trajName
- Can be specified per instrument to be displayed in the editor.
trajName
- specifies the trajectory file base name.
- The value is not explicitly exposed in the editor and should generally not be overwritten.
description
- metadata that describes trajectory.
- Is empty by default.
entryName
- defines the format for entries for NEXUS data files, by default is set to "" (an empty string).
- can be specified per instrument to be displayed in the editor.
- Default: empty string
fileGroup
- defines a trigger for incrementing fileNum.
- A list of evaluated fileGroups will be maintained per trajectory and reset every time a new one starts.
- By default, fileGroup is empty and does not change; therefore, fileNum increments only once per trajectory.
- SANS instruments can take advantage of this feature.
- In a typical trajectory, every point needs to write to a new data file. It can be accomplished by making fileGroup change at every point: fileGroup = pointNum.
- Can be specified per instrument to be displayed in the editor.
- Default: empty string
Default File Rules
The following default file rules are used if not explicitly defined in a trajectory:
fileName = filePrefix + fileNum
filePrefix = trajName
entryName = empty string
fileGroup = empty string
Whenever a new trajectory is created through the NICE editor, file rule expressions are explicitly set to default values as determined by the instrument scientist. However, it is still possible for these file rules to be undefined given that:
- Older trajectories may not have defined these expressions explicitly.
- A user may delete these expressions in the editor.
- A user may create a trajectory outside the NICE editor.

