Summary
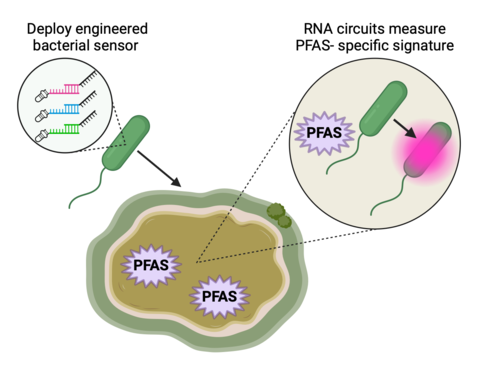
The Cellular Engineering Group is working to engineer components of living cells to sense environmental fluctuations induced by contaminants within their ecosystem. Living measurement systems can serve as real-time monitors of environmental contaminants like forever chemicals. More specifically, we seek to develop a cascade of RNA interactions to sense differential expression pattern changes induced by exposure to PFAS.
Description
Per- and polyfluoroalkyl substances, PFAS, are thermodynamically stable organic fluorinated chemicals that are resistant to traditional degradation pathways leading to concern for bioaccumulation. These compounds originate from several industrial manufacturing processes and are found in soil systems, food sources, and drinking water. Long-term consequences of PFAS to human health and environmental health are a growing concern. Therefore, detection and characterization of lasting effects on ecosystems and human health are paramount for remediation efforts.
Microorganisms are optimal living measurement systems to sense PFAS as they are ubiquitous in environments and naturally monitor and adapt to environmental fluctuations. We are leveraging RNA sequencing and transcriptomic analysis to measure and characterize changes in gene expression of microorganisms treated with PFAS. Cotranscriptionally encoded RNA strand displacement (ctRSD) circuits are programmed to recognize these identified differential expression patterns of interest and then genetically encoded into the microorganisms for real-time detection of PFAS induced RNA signatures.
The Cellular Engineering Group is part of the National Institute of Standards and Technology (NIST) Material Measurement Laboratory (MML) Biosystems and Biomaterials Division (BBD).
RElated Publications
- Wintenberg, M.E., Vasilyeva, O.B., Schaffter, S.W., Comparative transcriptomic analysis of perfluoroalkyl substances-induced responses of exponential and stationary phase Escherichia coli. BMC Genomics. 2025. DOI: https://link.springer.com/article/10.1186/s12864-025-12109-4

