Summary
RNA is an increasingly attractive molecule for engineering biology, in part due to the sequence-programmable nature of RNA:RNA interactions. The Cellular Engineering Group is developing predictable and versatile RNA circuits, based on nucleic acid strand displacement reactions, to meet emerging molecular computation and measurement needs in biological systems.
Description
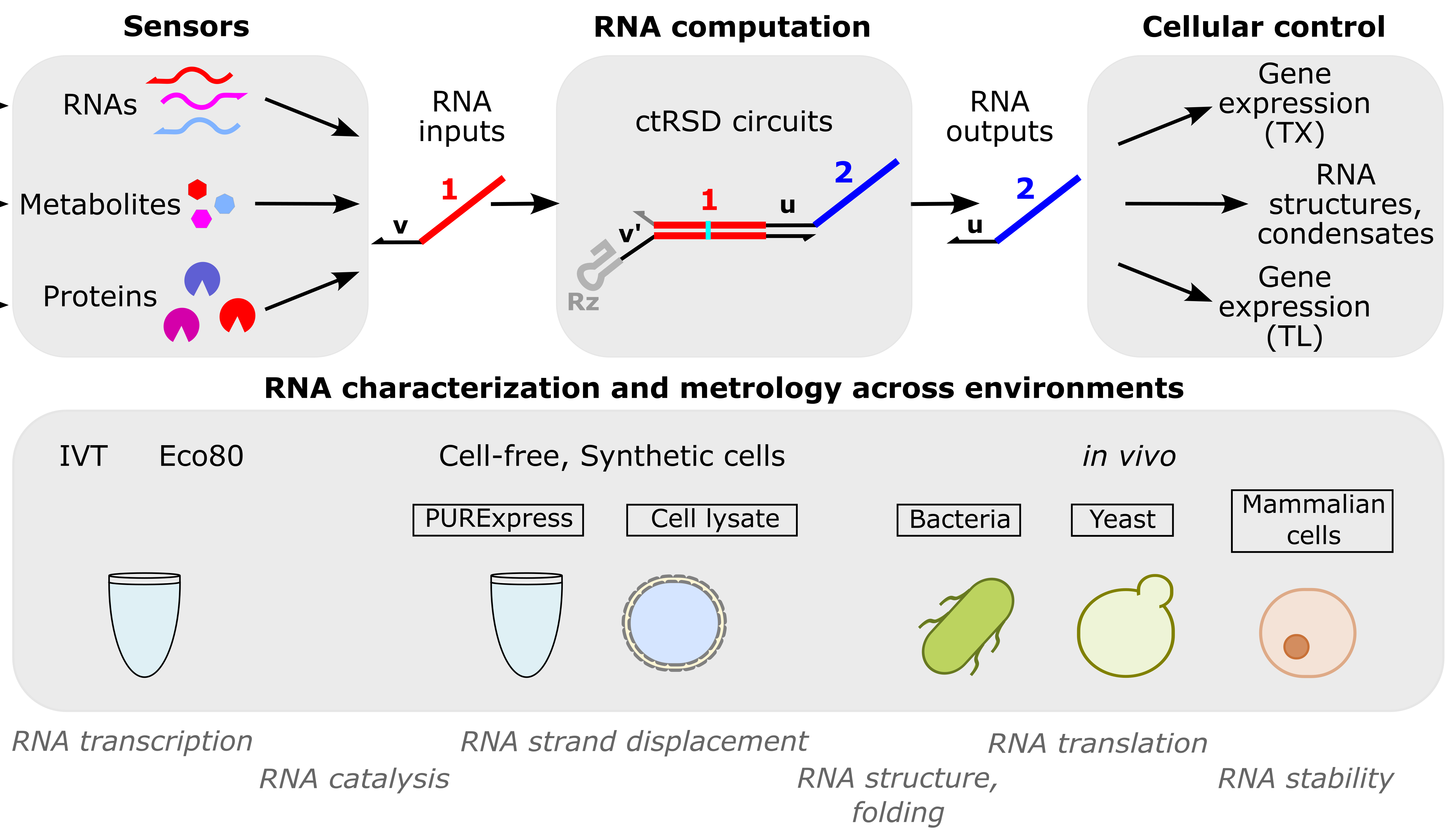
RNA computation
RNA computation, i.e., programmed interactions between RNA molecules that process information, is an increasingly useful synthetic biology tool with applications spanning in vitro biosensing, synthetic cells, and cellular control. We have developed cotranscriptionally encoded RNA strand displacement (ctRSD) circuits, which operate via toehold-mediated strand displacement. ctRSD circuits can be rationally programmed via sequence complementarity to execute logic, signal amplification, and complex information processing and decision making (Refs 3,4,5). Ultimately, ctRSD circuits can be genetically encoded for continuous RNA computation in living cells, cell lysates, or biological samples.
Sensors
RNAs: As RNA computation naturally operates on RNA inputs, it is well suited for measuring patterns of differential RNA expression. We use microbial RNA-seq measurements to identify differential RNA expression patterns unique to specific environmental stimuli and are programming RNA circuits to recognize these patterns in real-time inside living cells. This could enable a generalizable approach to engineering living measurement systems for any set of stimuli.
Metabolites: Allosteric transcription factors (aTFs) are routinely used to sense small molecules and metabolites in synthetic biology. As aTFs regulate transcription, they transduce metabolite binding to RNA expression for RNA computation. Leveraging large-scale measurements in the Living Measurement Systems Foundry (LMSF), we can precisely engineer aTF response and engineer new ligand specificities. These aTFs can be integrated with RNA computation for multi-input metabolite measurements.
Proteins: Riboswitches are typically used to transduce protein binding to a transcriptional response, but tuning response and engineering designs that work for new proteins remains challenging. In collaboration with Johns Hopkins University, we have developed a versatile protein biosensing platform, termed ARTIST, in which protein-aptamer binding regulates RNA transcription (Ref 2). ARTIST serves as a platform to transduce protein inputs to RNA inputs that can be processed with RNA computation.
Cellular control
A major goal of our RNA computation work is to engineer a cellular response when a specified set of molecular inputs is measured. For example, cell-based factories that dynamically optimize their yield in biomanufacturing or cells that diagnose themselves with cancer or other diseases as soon as the disease pattern arises. Towards this goal we are developing technologies to connect RNA computation outputs to cellular function, with a current emphasis on regulating protein expression.
RNA characterization and metrology across environments
RNA computation has applications across many environments and organisms, from cell-free diagnostics and biosensors to living measurement systems and therapeutics. To support RNA computation across these applications, we develop techniques to measure RNA processes in diverse environments (Ref 1). ctRSD circuits serve as a model system, but the measurements are applicable to any RNA-based technologies, such as riboswitches, CRISPR guide RNAs, small interfering RNAs, antisense oligonucleotides, and mRNA therapeutics. This work leverages the group’s expertise in cell-free expression systems.

RELATED PUBLICATIONS
- Alperovich, N.Y., Vasilyeva, O.B., Schaffter, S.W., Prevention of ribozyme catalysis through cDNA synthesis enables accurate RT-qPCR measurements of context-dependent ribozyme activity. bioRxiv. 2024. DOI: https://doi.org/10.1101/2024.07.19.604288
- Lee, H., Xie, T., Kang, B., Yu, X., Schaffter, S.W., Schulman, R. Plug-and-play protein biosensors using aptamer-regulated in vitro transcription. Nature Communications. 2024. DOI: https://doi.org/10.1038/s41467-024-51907-4
- Schaffter, S.W., Wintenberg, M.E., Murphy, T.M., Strychalski, E.A. Design approaches to expand the toolkit for building cotranscriptionally encoded RNA strand displacement circuits. ACS Synthetic Biology. 2023;12(5): 1546–1561.DOI: https://pubs.acs.org/doi/full/10.1021/acssynbio.3c00079
- Schaffter, S.W., Strychalski, E.A. Cotranscriptionally encoded RNA strand displacement circuits. Science Advances. 2022;8(12). DOI: https://www.science.org/doi/10.1126/sciadv.abl4354
- Schaffter, S. W.; Murphy, T. M. ctRSD_simulator_2.0. https://ctrsd-simulator.readthedocs.io/en/latest/
RELATED CONTENT
Sam Schaffter Presents at Build-A-Cell Seminar: https://www.youtube.com/watch?v=vSkwl4x6JP0
Sam Schaffter Interviewed on Meet the Molecular Programmer Podcast: https://podcast.molpi.gs/media/schaffter-s-1c8799780ad4af6f/

